The TSC Natural History Database captures clinical data provided by physicians to document the impact of the disease on a person’s health over his or her lifetime. The TSC Biosample Repository stores samples of blood, DNA, and tissues scientists can use in their research. The samples we collect are all linked to clinical data in the TSC Natural History Database.
More than 2,600 people with TSC are enrolled in the Natural History database and more than 1,000 people have contributed biosamples. The biosamples and data about individuals who provided samples help researchers discover biomarkers of TSC, test potential drug treatments, and determine why TSC is so different from person to person. If you are interested in participating or learning more, please fill out the interest form at the bottom of the page.
Testimonials
“A blood draw is such an easy thing for my daughter to do. To have it done at home makes it super convenient.”
“We want to see a cure in our lifetime and have to say things are so much better than 30 years ago because of the efforts of researchers and the TSC Alliance.”
“By participating in the Tuberous Sclerosis Complex Research and the Biosample Repository Program, we know we will bring new hope for patients afflicted, no matter how immediate.”
“Our family chose to participate in this initiative because we have seen the progress made over our son’s lifetime due to research advances.”
“The TSC Alliance’s mobile phlebotomy initiative made it easy for our daughter to participate safely and in the comfort of our own home.”
“I chose to participate in the Biosample Repository because I was asked by caring people in similar situations and it is the right thing to do for her.”
“We are so grateful for the many advances in TSC Research and for the patients and parents who have offered data and samples, contributing to this great progress.”
“Our hope is that by participating maybe we will benefit from the research but if not, others will.”
“We highly recommend any parent or TSC adult to participate in the process to seek cure of TSC.”
Biosample contribution
Blood samples
The TSC Biosample Repository is collecting samples from TSC Natural History Database participants at specific clinics and via mobile blood collections. In coordination with a national mobile phlebotomy service (TravaLab or ExamOne), the TSC Alliance can collect blood samples from anyone in the United States with TSC. If you are interested in participating or learning more, please fill out the brief interest form at the bottom of the page. Someone from the Science and Medical team will be in touch shortly. You can also let us know your interest by emailing biosample@tscalliance.org. The TSC Alliance hopes to collect blood samples from participants once per year.
The TSC Alliance also accepts blood samples from volunteers enrolled in the TSC Natural History Database at the following sites:
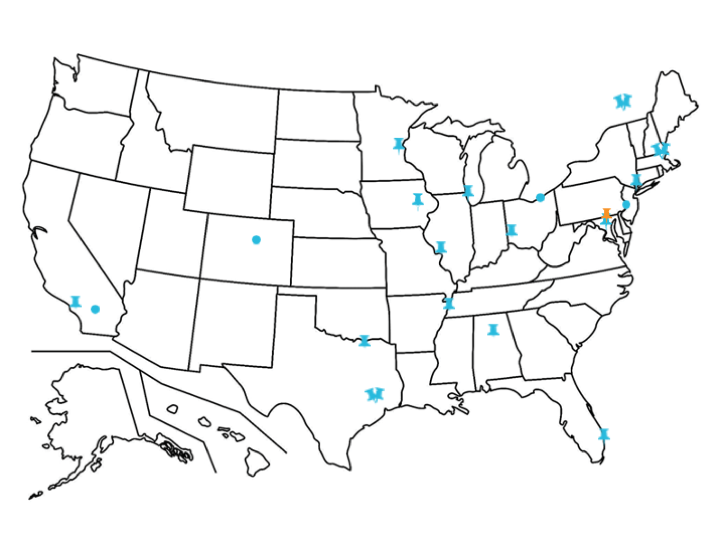
Circle icons indicate a database only site meaning that biosamples are not collected on site. Pushpins indicate sites approved to collect biosamples on site
Individuals with TSC seen at one of the participating institutions may ask their health care provider or clinic coordinator about participating in the TSC Natural History Database and Biosample Repository. If you are interested in participating or learning more, please fill out the brief interest form at the bottom of the page.
Research updates
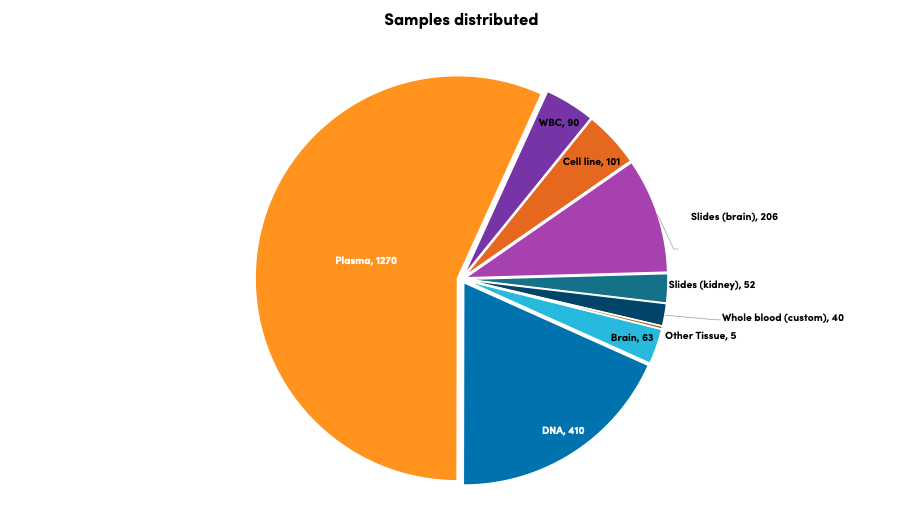
Biosample distribution
Since inception, the TSC Biosample Repository has distributed 2,237 samples to 47 distinct researchers for 56 distinct projects. Some examples of projects conducted using biosamples include bioassays measuring levels of cellular activity, screening for novel therapeutic targets, detecting biomarkers, and measuring metabolites in blood and comparing them with phenotypes reported in the TSC Natural History Database. DNA extracted from cheek swabs and blood samples can be used for genetic testing to detect TSC variants, and plasma from blood samples can be used to assay for predictive biomarkers or biomarkers of response and tissue samples are used to detect tissue-specific markers and cell types that are important for studying TSC progression.
For researchers
If you are a researcher interested in using samples from the TSC Biosample Repository in your work, please see here for more information.
Post-mortem brain donation
The University of Maryland Brain and Tissue Bank (MBTB) makes arrangements for this type of precious gift on behalf of the TSC Alliance. Individuals living anywhere in the United States who wish to donate whole brain tissue after death should contact MBTB at btbumab@som.umaryland.edu. If death is imminent and you would like to donate brain tissue, please call the MBTB as soon as possible at 1-800-847-1539.
Successful donation rests on swift and thorough communication between family members, healthcare professionals, and the MBTB. Although the MBTB will make every effort to retrieve tissue in an emergency, tissue recovery may be impossible if there is no advance notice. Please click here for instructions on how to register in advance for postmortem donation. There is no cost to the family to donate.
Steven Sparagana Legacy Fund
The TSC Natural History Database and Biosample Repository are funded by donors who contribute to funds such as the Steven Sparagana Legacy Fund, which provides an ongoing source of revenue in support of the Natural History Database and Biosample Repository as a tribute to Dr. Sparagana’s contributions to the creation and ongoing success of these invaluable tools. Learn more about the fund and donate here.
Acknowledgments
The TSC Natural History Database and Biosample Repository are governed and wholly funded by the TSC Alliance thanks to generous support from Lorne Waxlax, William Watts, Dr. Michael and Janie Frost, Janice and Julian Gangolli Family Fund, Jim and the late Andrea Maginn, and many additional donors through the Unlock the Cure campaign.
Our thanks to UCB, Inc. for supporting translation of consent forms for this project into Spanish.
Interested in participating in the TSC Natural History Database and Biosample Repository Project?
Fill out the brief interest form below and a TSC Alliance Science and Medical team member will be in touch with you shortly.